Core-hole localization#
A special aspect of X-ray spectroscopy calculations which involve an explicit core-hole, e.g. X-ray photoelectron spectroscopy (XPS) or X-ray emission spectroscopy (XES), is the delocalization-induced relaxation error (DIRE) [BF22]. DIRE affects symmetric molecules where care must be taken to localize the core-hole, especially when HF or DFT theory levels are used. Below follows an illustration of DIRE in the case of the ethylene molecule. We will calculate the ionization energy of the C 1s core orbitals. The ionization energy is calculated as:
We will have to perform a ground state calculation, as well as a calculation with a full core-hole (FCH) on the C 1s orbital. The FCH is obtained using the maximum overlap method (MOM). One option to localize the core-hole is to replace the 1s electrons of the non-core-ionized C atom with an effective core potential (ECP). Another option is to very slightly distort the molecule, which is the approach adopted here.
Step 1 \(-\) Define the molecular geometries#
In the following, we will create two ethylene molecules, one fully symmetric to obtain delocalized C 1s core orbitals and and one with slightly stretched CH bonds at one of the C sites, to obtain localized C 1s core orbitals.
deloc_ethylene_xyz = """6
symmetric ehtylene molecule
C 0.666005 0.000000 0.000000
C -0.666005 0.000000 0.000000
H 1.227874 0.922836 0.000000
H 1.227874 -0.922836 0.000000
H -1.227874 0.922836 0.000000
H -1.227874 -0.922836 0.000000
"""
loc_ethylene_xyz = """6
asymmetric ethylene molecule to localize the C1s orbitals
C 0.666005 0.000000 0.000000
C -0.666005 0.000000 0.000000
H 1.227874 0.922836 0.000000
H 1.227874 -0.922836 0.000000
H -1.2538762 0.965543 0.000000
H -1.2538762 -0.965543 0.000000
"""
basis_set_label = "6-311++G_D,P_" #"DEF2-TZVPPD" #"TAUG-CC-PCVDZ" #
deloc_ethylene = vlx.Molecule.from_xyz_string(deloc_ethylene_xyz)
loc_ethylene = vlx.Molecule.from_xyz_string(loc_ethylene_xyz)
basis = vlx.MolecularBasis.read(loc_ethylene, basis_set_label)
Step 2 \(-\) determine the ground state energy#
deloc_scf_drv = vlx.ScfRestrictedDriver()
deloc_scf_drv.ostream.mute()
deloc_scf_results = deloc_scf_drv.compute(deloc_ethylene, basis)
loc_scf_drv = vlx.ScfRestrictedDriver()
loc_scf_drv.ostream.mute()
loc_scf_results = loc_scf_drv.compute(loc_ethylene, basis)
plot_orbital(deloc_ethylene, basis, deloc_scf_drv, orbital_index=0)
plot_orbital(loc_ethylene, basis, loc_scf_drv, orbital_index=0)
Step 3 \(-\) determine the core-ionized state energy#
def compute_fch_energy(molecule, basis, scf_drv):
fch_molecule = copy.deepcopy(molecule)
fch_molecule.set_charge(1)
fch_molecule.set_multiplicity(2)
# Lists of occupied alpha and beta MOs
occ_alpha = [0, 1, 2, 3, 4, 5, 6, 7]
occ_beta = [1, 2, 3, 4, 5, 6, 7]
# Calculate SCF of the core-ionized system using the maximum overlap method
scfdrv_fch = vlx.ScfUnrestrictedDriver()
scfdrv_fch.ostream.mute()
scfdrv_fch.maximum_overlap(fch_molecule, basis, scf_drv.mol_orbs,
occ_alpha, occ_beta)
scf_results = scfdrv_fch.compute(fch_molecule, basis)
return scfdrv_fch.get_scf_energy()
deloc_fch_energy = compute_fch_energy(deloc_ethylene, basis, deloc_scf_drv)
deloc_gs_energy = deloc_scf_drv.get_scf_energy()
deloc_hf_ie = (deloc_fch_energy - deloc_gs_energy) * hartree_in_ev()
# Add a Lorentzian broadening
deloc_x, deloc_y = lorentzian(np.array([deloc_hf_ie]), np.array([1]),
deloc_hf_ie - 5, deloc_hf_ie + 5, 0.01, 0.7)
loc_fch_energy = compute_fch_energy(loc_ethylene, basis, loc_scf_drv)
loc_gs_energy = loc_scf_drv.get_scf_energy()
loc_hf_ie = (loc_fch_energy - loc_gs_energy) * hartree_in_ev()
loc_x, loc_y = lorentzian(np.array([loc_hf_ie]), np.array([1]),
loc_hf_ie - 5, loc_hf_ie + 5, 0.01, 0.7)
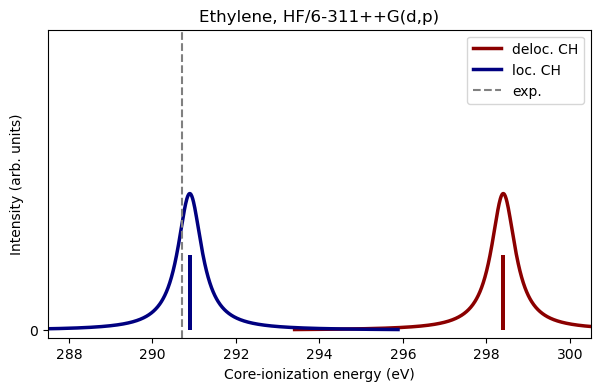
Now that we have calculated the ionization energies for both the localized and delocalized CH, let us plot the results. The dotted line marks the measured ionization energy [HBorveSaethreT11].
plt.figure(figsize=(7, 4))
plt.bar(deloc_hf_ie, 1.0, color="darkred", width=0.1)
plt.bar(loc_hf_ie, 1.0, color="navy", width=0.1)
plt.plot(deloc_x, deloc_y, color="darkred", label="deloc. CH", linewidth=2.5)
plt.plot(loc_x, loc_y, color="navy", label="loc. CH", linewidth=2.5)
plt.xlabel("Core-ionization energy (eV)")
plt.ylabel("Intensity (arb. units)")
plt.title("Ethylene, HF/6-311++G(d,p)")
plt.axvline(290.7, linestyle="--", color="gray", label="exp.")
plt.axis(xmin=287.5, xmax=300.5, ymin=-0.1, ymax=4.0)
plt.xticks([288, 290, 292, 294, 296, 298, 300])
plt.yticks([0])
plt.legend()
plt.show()
Show code cell source
print("DIRE, HF XPS: %.2f eV" % (deloc_hf_ie - loc_hf_ie) )
DIRE, HF XPS: 7.51 eV
